
|
Dinosolve: a protein disulfide bonding prediction server
|
 |
|
C3-Scorpion: a protein 3-state
secondary strcutre prediction server |
 |
|
C8-Scorpion: a protein 8-state
secondary strcutre prediction server |
 |
|
Casa: a protein 2-state
Solvent Accessibility
prediction server |
 |
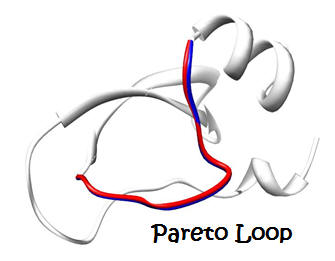 |
|
|
ICOSA: A Distance-dependent, Orientation-specific
Coarse-grain Contact Potential for Protein Structure
Modeling |
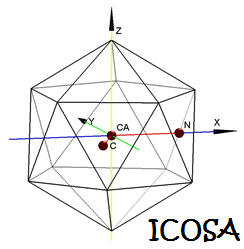 |
|
Phosphorylation Sites Prediction |
 |